Anticancer area:
Our primary interest in the anticancer area is to investigate the cellular processes of human DNA replication and repair. We study proteins that are involved in both DNA replication and Base excision repair (BER) and Nucleotide exchange repair (NER) processes. The proper functioning of proteins involved in these processes is essential for maintaining genomic integrity and any problems with their function, expression or interactions can lead to human diseases such as cancer. The importance of DNA repair proteins has been recently highlighted with the 2015 Nobel prize for Chemistry going to Paul L. Mordick, Tomas Lindahl and Aziz Sankar in the field of DNA repair.
Our lab is studying DNA replication proteins such as Polδ, PCNA, RFC, Fen1, Lig1, PARP1 and Topo1 and their interactions with other proteins. Polδ is primarily involved in the synthesis and processing of the lagging strand DNA (composed of Okazaki fragments). PCNA is a beta clamp that harbours Polδ on the DNA. RFC is a clamp loader that loads PCNA onto the DNA, while Lig1 seals the gaps between Okazaki fragments to make the continuous lagging strand DNA. The interplay among these proteins, their post-translational modifications, mutations or other factors that lead to changes in their function, expression or interaction makes for very interesting studies with implications for genomic instability and cancer
Lig1 and FEN1 have been recently described as biomarker for breast and ovarian cancers. Clinical trials for PARP1 and Topo1 inhibitors for cancer were also in the news recently. Taking cue from these, we have conducted a pharmacophore based screening for hLig1 and Fen1 inhibitors from the CDRI compound library and have found hLig1 inhibitors that can slow down the progression of breast cancer in a mouse model. Fen1 inhibitors are being currently tested for their anticancer properties. Our lab has also developed assays for Topo1 and PARP1 and inhibitors for these proteins will be screened shortly.
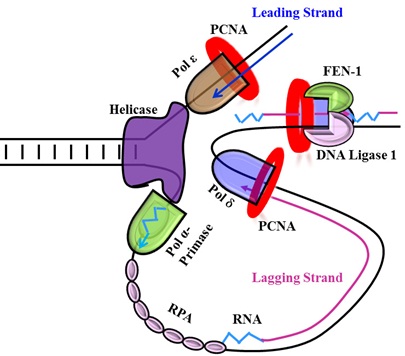
Characterization of DNA replication proteins involved in lagging strand synthesis and maturation
In eukaryotes, the process of cell division is a complex phenomenon and is preceded by the division of the nuclear DNA. Nuclear DNA replication is carried out by a network of proteins and enzymes that work rapidly in a well-coordinated manner to duplicate the all-important genetic information. Several components of the replication apparatus perform their exact roles in a complex called the replisome that are located in so called nuclear “replication factories”. For replication to begin the chromosomes must unwind and the synthesis of both new strands must occur simultaneously.
In the context of nuclear DNA replication, the process of lagging strand synthesis is far more dymanic and complex than the leading strand synthesis and includes under its ambit a larger number of proteins and cofactors. Studies on DNA replication and repair have revealed that three important accessory factors along with the DNA polymerase δ (Pol δ) are needed for successful replication of the lagging strand. One is a homo-trimeric DNA clamp protein called PCNA (Proliferating cell nuclear antigen); the second is a hetero pentameric RFC (Replication factor C) protein and the third is a hetro-trimer called RPA (Replication protein A). Other than these the ligase I enzyme is involved in the maturation of Okazaki fragments. Post-translational modifications (PTMs) are also known to affect lagging strand synthesis.
Our lab works on replication proteins PCNA, RFC, Pol δ, LigI and Fen1. Some questions we are trying to answer are:
Approximately 10–15% of patients with breast cancer has an aggressive disease and develops distant metastases within 3 years after the initial detection of the primary tumor. The heterogeneous nature of breast cancer metastasis makes it difficult not only to define cure for this disease, but also to assess risk factors for metastasis. Owing to their invasive potential, it is very likely that they manipulate the protein turnover of key regulatory proteins involved in tumor regression. As protein turnover is maintained by E3s and DUBs, we hypothesize that identification deregulated E3s, DUBs and their substrates may be pivotal to understanding the pathophysiology of breast cancer invasion and better therapeutics.
How does the interaction among these proteins affect the process of strand synthesis?
How these proteins are targeted to the replication factory?
We employ a combination of cell biology, molecular biology and biophysical techniques to elucidate the role of these proteins and their interaction in DNA strand synthesis
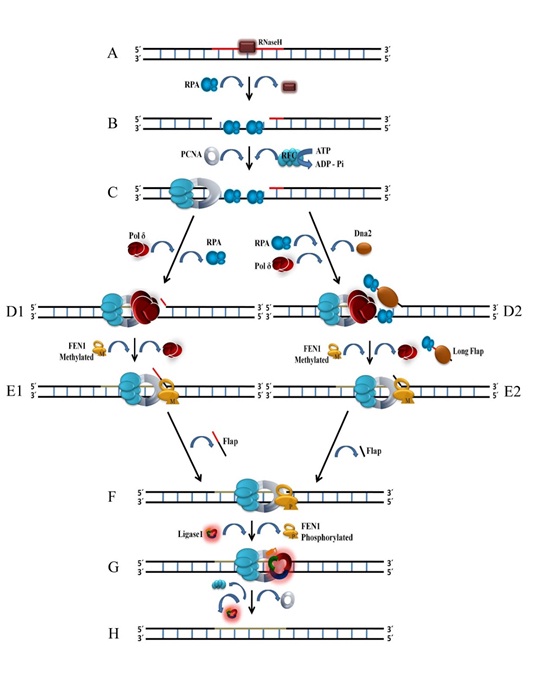
Figure: Cartoon depicting the stepwise involvement of proteins in lagging strand DNA synthesis.
Human DNA ligases: Novel targets for the development of new anticancer drugs
In humans, three types of DNA ligases (human DNA ligase I, III, IV/XRCC4) are found. Human DNA ligase I (hLigI) is mainly involved in the joining of nascent DNA (Okazaki fragments) generated during the process of replication and is also involved in DNA repair pathways like long patch base excision repair pathway (single strand break repair pathway) and micro-homology mediated end-joining (MHEJ) pathway (double strand break repair pathway). Human DNA ligase III (hLigIII) is involved in single strand break repair (SSBR) pathways like short patch base excision repair (BER) pathway and nucleotide excision repair (NER) pathway. Human DNA ligase IV (hLigIV/XRCC4) performs DNA nick sealing in double strand break repair pathway (DSBR) by non-homologous end joining (NHEJ). Looking at the functions performed by DNA ligases, it seems that they play very significant roles in maintaining genomic integrity. Our lab focuses on hLigI inhibitors because it is important for DNA replication and is also shown to be over-expressed in some rapidly dividing cancer cells. Normal non-replicating cells express hLigI at very low levels. Therefore, the inherent targeting of hLigI inhibitors to the cancer cells can promote cellular apoptosis leading to arrest of cancer progression and at the same time causing much less toxicity to normal cells than non-targeted chemotherapies.
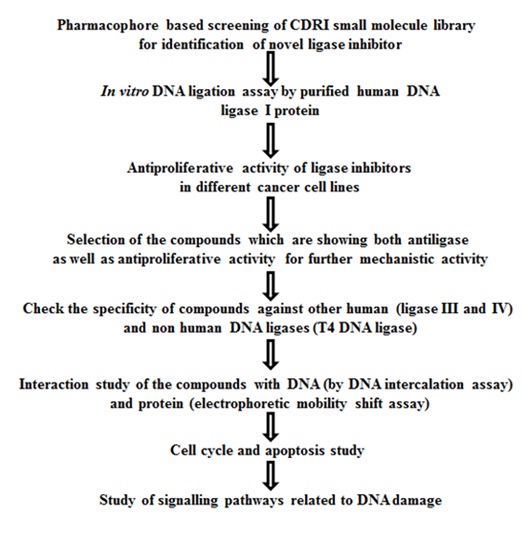
Figure: Cartoon depicting the stepwise involvement of proteins in lagging strand DNA synthesis.
Reversal of drug resistance in the human pathogenic fungus Candida albicans.
Functional Characterization and Identification of Small Molecules Inhibitors for TAC1, a transcriptional activator of CDR genes in Candida albicans.
Candida albicans is a major cause of opportunistic and nosocomial infections in humans all over the world. The last two decades have seen the emergence of resistance of Candida sp. to the commonly used azole antifungal drugs. This is mainly due to up-regulation of ABC transporters CDR1 and CDR/2. TAC1 has been identified as the transcriptional activator of CDR1/2 genes and has no human homologues. Therefore, studies to understand the molecular regulation of ABC transporter by TAC1 are being performed in the laboratory. Inhibitors against Tac1 are being screened using in silico pharmacophore based and docking approaches. In vitro and in vivo testing of small molecule inhibitors Tac1 for their ability to sensitize drug resistant clinical isolates of C. albicans to azole and other antifungal drugs is being done.
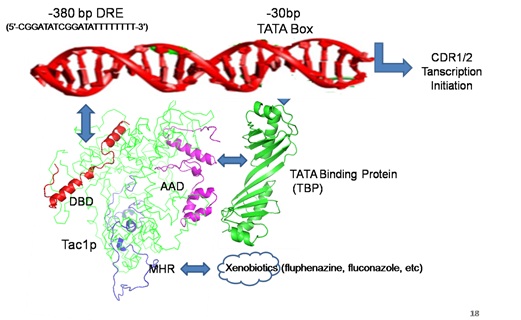
Figure: Cartoon showing TAC1 binding to the promoter of ABC transporter genes CDR1/2 of Candida albicans. Blocking TAC1 interaction can lead to downregulation of ABC transporters as a novel strategy for reversal of drug resistance.