X-ray lab (Prof. G.N. Ramachandran Structural Biology laboratory)
Crystal structure information is invaluable for modern drug discovery. CDRI has a team of structural biologists who structurally and functionally characterise protein targets from human pathogens that cause infectious diseases like leishmaniasis, tuberculosis and filariasis that disproportionately affect disadvantaged sections of our society. The team also works on highly specialized research areas such as in-situ cryo-crystallography, high resolution experimental charge density studies, polymorph screening and development of pharmaceutical salts/co-crystals based crystalline pre-formulations of drugs.
Structures of proteins/ macromolecular targets are used in conjunction with rational strategies to identify new inhibitors. Co-crystal structures with initial hits then forms the basis for medicinal chemists to optimise the potency and positive properties of new lead with therapeutic potential. Structures of small molecules helps medicinal chemists to unambiguously characterise the newly synthesized compounds.

290 K
|
|

265 K
|
|

260 K
|
| Snapshots of in-situ cryo-crystallization of liquid sample.
|
| |
|
|
|
|
|
|
|
|
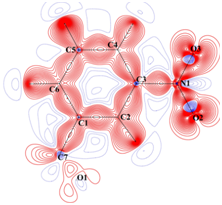
Static deformation density
|
|
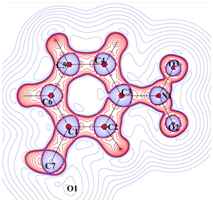
Laplacian of electron density
|
|
Charge density distribution determined from high resolution (~0.45 Å)
X-ray diffraction studies. |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
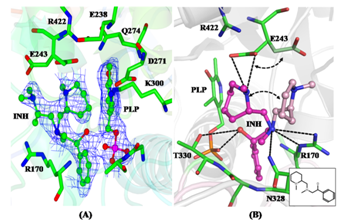
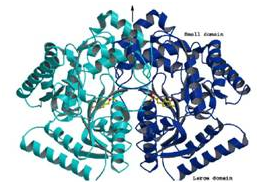
| (A) 2Fo-Fc electron density map contoured around a 2-aminomethyl piperidine derivative identified as an inhibitor of M. tuberculosis Lysine-ε-aminotransferase (LAT). (B) Protein-inhibitor interactions: The inset shows the 2D representation of the inhibitor. (C) Crystal structure of LAT
|
|
|
|
|
| |
|
|
|
|
|
|
|
|
|
|
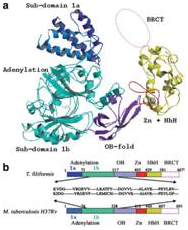
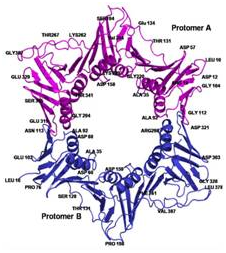
(From Left-to-Right) Crystal structures of a mutant NAD+-dependent
DNA ligase (LigA) from M. tuberculosis, Sliding DNA β-clamp (DnaN), and
identification of specific anti-LigA inhibitors.
|
| |
|
|
|
|
|
|
|
|
Static deformation density
|
|
Laplacian of electron density
|
|
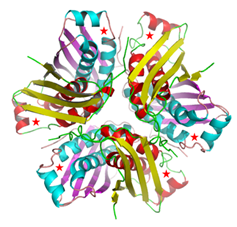
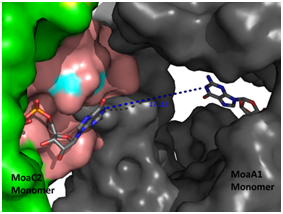
Crystal Structure of the M. tuberculosis MoaC (Rv0864) hexamer
(Left panel), asteriks denote the ligand binding site.
(Right panel) Computational docking of MoaC with MoaA (grey), enzymes performing
successive steps in the Molybdenum cofactor biosynthesis pathway, showing the
channel that connects the ligand binding sites of both the enzymes .
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
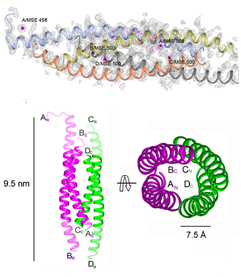
Crystal Structure of L. donovani coronin coiled coil domain,
that assembles as an anti-parallel tetramer (Bottom left). The structure
was solved by Se- SAD and their positions are shown (Left, top).
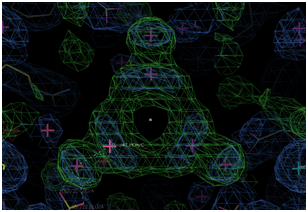
(Right Panel) Electron Density of a water cluster, as viewed down the
crystallographic 3-fold symmetry axis in another structure.
|
|
|
|
|
The state-of-the-art facility in the institute includes
X-ray generators and detectors to collect data and solve structures of
macromolecules/ proteins and small-molecules:
-
➤MOSQUITO liquid handling robotic system for setting up protein crystallization experiments
-
➤CRYSTALPRO imaging system with a 28 plate hotel for automated image analysis of protein crystallization plates
-
➤RIGAKU Micromax-007HF X-ray generator, Varimax optics, MAR345-DTB image plate system, OXFORD 700 series cryocooler to collect data from protein/macromolecule crystals.
-
➤RIGAKU dual source Kappa diffractometer equipped with an AFC12 goniometer and enhanced sensitivity (HG) Saturn724+ CCD detector. An OXFORD 700 series cryocooling system helps with variable temperature studies involving small-molecules
-
➤Computational servers and workstations to conduct complex calculations and graphics visualisation.
The highly qualified and experienced team offers consultancy and collaborations
with pharmaceutical companies under transparent time-bound MOU and mutually
agreed terms.
Please contact: Dr. R. Ravishankar email:
r_ravishankar@cdri.res.in